A discovery in Marina Guenza’s UO chemistry lab, published in a major physics journal, is already being tapped by outside scientists working on a new medical treatment for tuberculosis.
The UO research, however, included a detour. Jeremy Copperman, a doctoral student leading it, was diagnosed with Hodgkin's lymphoma in the final year of his doctoral studies in physics. His work slowed amid six months of chemotherapy.
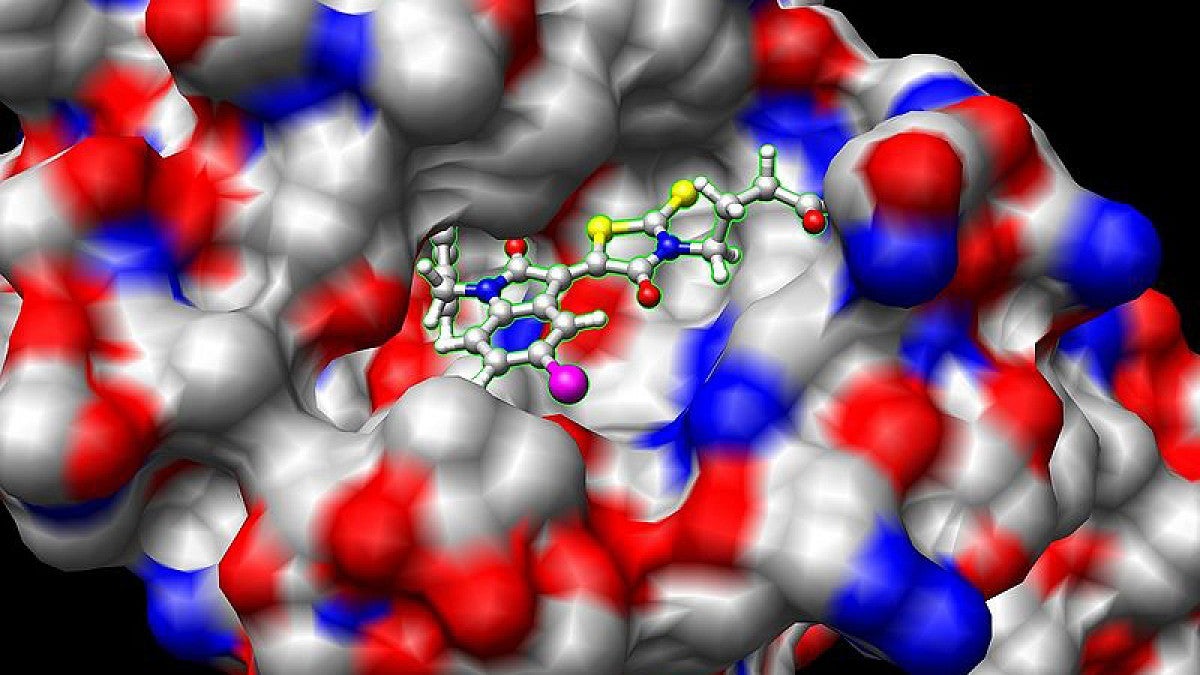
In the paper, the trio and Guenza, a professor in the Department of Chemistry and Biochemistry and member of the Institute of Theoretical Science, reported that biological proteins move in a universal way. In a narrow window of conditions where a protein performs its biological function, the motion critically slows in a universal fashion that can be mapped into an equation for analysis.
Proteins are vital workhorses in biological systems. When something goes wrong, they can fuel disease. The study has immediate ramifications, Guenza said, because it identifies when a protein is open to binding with other proteins or to accept a small molecule such as a drug.
The National Science Foundation-supported work, even before publication, swiftly drew the interest of Andrzej Joachimiak of the University of Chicago and director of two labs at the Argonne National Laboratory. His multi-institutional team recently announced the discovery of a molecule that knocks out a protein vital to the survival of the bacterium that causes tuberculosis.
Joachimiak’s group knows its molecule binds to the targeted protein, but not how and where. The approach in Guenza’s lab, which he heard about from a presentation Guenza gave at the Second International Conference on Computational Genomics and Proteomics in Panama, will be used in a collaborative effort to answer those questions. If successful, the new drug could be tweaked to make it more effective.
For Copperman, who grew up in Eugene and had earlier earned a bachelor’s degree in physics from the UO, the paper’s publication provided a gratifying closing chapter to his work at the UO.
He and his wife, Erika, had owned and operated Three Forks Wok & Grill, a popular Eugene restaurant with two locations. They closed the business so he could focus on his doctoral research and she could pursue a medical degree.
After earning his doctorate in 2016, Copperman joined the University of Wisconsin–Milwaukee. His wife is now doing a residency in obstetrics and gynecology at a Milwaukee hospital.
At the UO, Copperman was intrigued by “the universality that ties together systems that on the surface look completely different.” Guenza, his advisor, tasked him to explore that idea on protein dynamics.
“I was modeling the dynamics of a protein called ubiquitin, and in the process of doing this, I noticed a trend, a scaling pattern, which had no reason to be there,” Copperman said. “I simulated the protein motion on the local supercomputer and spent weeks writing analysis codes. I repeated this process for six other proteins and found the same simple scaling pattern.”
As a next step, Copperman said, he theorized that the scaling was generated by an “energetic disorder in the time domain.” That, he said, includes small, random variations that become dramatic at increasing scales, similar to the front of an expanding wildfire.
That take drew curiosity and some skepticism from Guenza, who challenged Copperman to find “a stronger physical argument.” Eventually, he said, she agreed that the idea was compelling and decided to push it forward.
“This illustrates the great scientific relationship Marina and I had,” Copperman said. “With her physical intuition and healthy skepticism, together we continually improved the idea.”
After Copperman left the UO, Dinpajooh and Beyerle kept working, spending months simulating and performing a more comprehensive analysis of a more extended set of proteins using a supercomputer at the University of California, San Diego.
“The original idea for this research came from Jeremy,” Guenza said. “Without him, my lab never would have explored this interesting analogy between protein dynamics and motion in random disorder. And without Hadi and Eric, we would not have had this paper published.”
—By Jim Barlow, University Communications