One of the most widely used drugs to fight cancer could become more effective and safer, based on findings emerging from the lab of UO chemist Vickie DeRose.
In two recently published papers, separate projects using different approaches and led by two of DeRose’s doctoral students have made new predictions for where the platinum-carrying drug cisplatin binds in cells.
While used for more than 50 years in frontline treatments against several types of cancers, there has been little concrete information on how cisplatin and its derivatives work, DeRose said. Their use also can generate a long list of undesired side effects, including serious kidney problems and nerve damage. Resistance to the drug also occurs.
When injected into a patient, the drug goes everywhere, finding its way to both cancerous and healthy cells. Researchers have known the drug finds its way to DNA, RNA and proteins, but little about what happens when it gets there.
“We don’t know exactly how these compounds work in actually killing cancer cells,” DeRose said. “Some pathways have been defined, but a lot of information is not known. We don’t know what all the compounds bind to in the cells. We are trying to fill that hole in the knowledge base.”
The two recent studies, both of which relied on high-throughput techniques with yeast cells, shed new light.
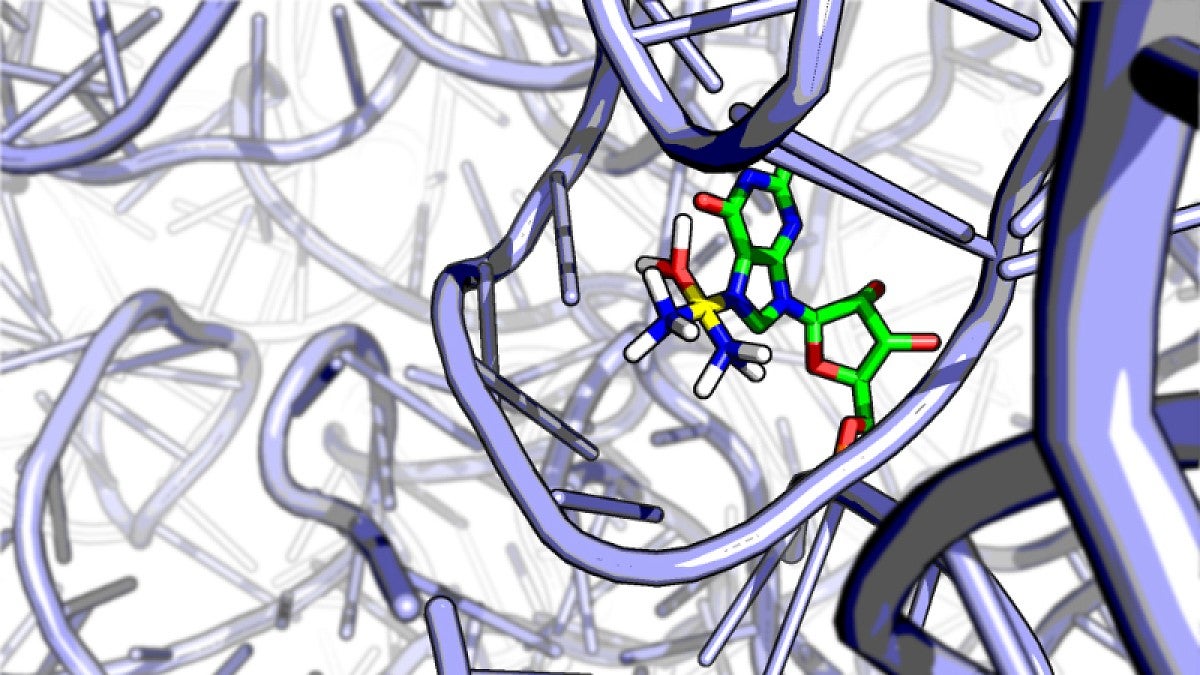
Using high-throughput sequencing and bioinformatics analysis — methods common in DNA studies but rare for spotting drugs on RNA — Kory Plakos discovered that cisplatin latches onto weak spots, tiny crevices, on ribosomal RNA, the cellular workhorse that carries out the instructions of DNA.
His findings were detailed in Chemical Communications, a journal of the U.K.’s Royal Society of Chemistry. His research builds on a discovery in 2011, in which DeRose documented that cisplatin binds to RNA up to 20-fold more than it does to DNA.
“We treated yeast cells with platinum, extracted the RNA and did a chemical process called a reverse-transcription assay,” Plakos said. “This process stalls at sites where platinum sits on a strand of RNA, allowing us to identify all of the potential binding sites that we can access. It seems to bind at weaknesses that are also targets of antibiotics. We found evidence that cisplatin interferes with the way the ribosome works.”
The ribosome is a sphere-shaped structure in cells that contains RNA and where protein synthesis occurs.
In the other study, published in ACS Chemical Biology, a journal of the American Chemical Society, Rachael Cunningham used a click-chemistry process and new platinum molecules developed in collaboration with Michael Haley’s UO lab, in which derivatives of cisplatin carry a small “tail” for tagging.
Cunningham used the molecular tail to pull out platinum-bound targets in yeast, isolating 152 proteins. Seven are linked to a cellular condition known as endoplasmic reticulum stress, which affects protein folding and is a recognized consequence of cisplatin treatment. One protein that is crucial to the protein-folding process also happens to carry platinum-loving amino acids in its active site.
“If the cell can’t adjust to this stress, the cell dies,” said Cunningham, who predicts that platinum might block this crucial protein’s activity. “I tested this in a test tube, looking to see if the protein’s enzymatic function was affected in any way. It was inhibited, indicating that platinum not only binds to this protein but it also inhibits its function. This is really interesting and needs to be further tested. This might shed light on why cisplatin kicks off this stress response.”
As the two projects were being done, DeRose said, she was intrigued at the emerging discoveries. She was a co-author on both papers.
“These two studies provide a very unbiased view of all the targets of these platinum compounds in cells,” DeRose said. “In Rachael’s case, we’re talking 152 proteins out of a possible 6,000 in this yeast proteome, and in Kory’s case the localization in the ribosome where he looked contains about 2,000 nucleotides, but he narrowed it down to just five sites that seemed to the most-likely binding sites.”
Yeast as a research model offers simplicity. It is a eukaryote, an organism with many cellular functions similar to those in humans. The next step is looking at human cancer cells, DeRose said.
“The fact that we don’t really have an understanding of what this widely used class of drug is doing is surprising, so there is a lot of room for improvement if we understand how it is affecting both unhealthy and healthy cells,” Plakos said. “Eventually, it might be possible to narrow cisplatin’s action to reduce off-target effects.”
Cunningham was supported by the American Heart Association. Plakos was supported by a National Institutes of Health Training Grant in Molecular Biophysics. Both also participated in outreach activities including the UO Content in Context SuperLessons K-12 program. National Science Foundation grants to DeRose also funded the research.
—By Jim Barlow, University Communications