Probiotics may be better in the future based on what UO researchers have seen in real time in the guts of live zebrafish.
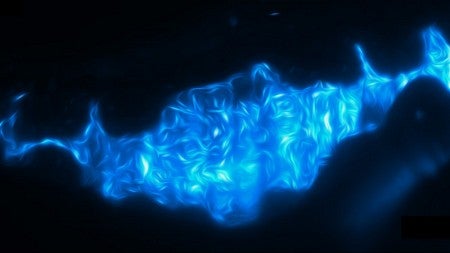
Numerous diseases are tied to gut microbiota. The probiotics industry targets intestinal health with products built mostly around enzyme cultures and bacteria. The UO discovery could lead to better products, said Raghuveer Parthasarathy, a professor of physics.
"This research shows that the physical environment and activity of the host intestine can be a major determinant of the bacterial makeup of the gut — an idea that we think should very generally influence how we tackle issues of health and disease,” he said.
Most studies have focused on the composition of microbial communities. Not knowing how bacteria are organized and how their populations change over time has kept scientists from understanding the mechanisms at play, he added. Diagnostic approaches currently capture only a moment in time and reveal nothing about spatial structure.
Parthasarathy likens that gap to trying to comprehend the ecology of the African savannah without knowing which organisms are stationary and which are mobile, and how the species interact.
AUDIO: Parthasarathy on his analogy to understanding life on the African savannah
Initially, UO researchers used non-imaging-based techniques to learn that two bacterial species native to zebrafish — Aeromonas veronii and Vibrio cholerae — have different abundances in the larval zebrafish gut. When zebrafish with intestines initially colonized by Aeromonas were introduced to Vibrio, Aeromonas populations shrank.
A conventional interpretation, Parthasarathy said, suggests a reduced growth rate of Aeromonas in the presence of Vibrio, but the researchers dug deeper.
They turned to a high-resolution, 3-D technique that was customized in 2014 in Parthasarathy's lab to peer inside zebrafish. Zebrafish larvae containing the two bacterial species were imaged every 20 minutes for 14 hours to get a precise map of bacterial populations in the gut. Researchers saw dramatic collapses of Aeromonas, followed by rebounding growth rates seen in bacteria on their own.
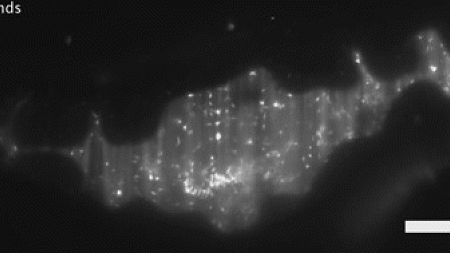
The researchers then looked at the two bacterial species in zebrafish carrying a mutation that leads to the loss of the brain-like network in the tissue lining of the gastrointestinal tract that orchestrates peristalsis. In the absence of peristalsis, the bacteria co-existed without competition.
This mutation is typical of human patients with Hirschsprung's disease, a birth defect that affects nerve cells in the large intestine and muscles involved in peristalsis. Usually seen in babies soon after birth, it leads to trapped fecal matter, infection and bowel problems.
The study opens a new window on conditions tied to problematic peristalsis, said Judith S. Eisen, a professor of biology and member of the UO Institute of Neuroscience.
"Zebrafish is really the only model in which these types of interactions can be visualized and studied in real time within the animal," she said. "Our work provides new insights into these types of host-microbe interactions and may also prompt new thinking about therapeutic approaches for human disease."
Eisen and Karen Guillemin, a professor of biology, noted that impaired peristalsis might profoundly affect gut microbiota by altering competition.
"Microbiome-profiling studies of Hirschsprung's patients provide evidence that their microbiomes are different from healthy controls," Guillemin said. "About 30 percent of these patients develop what is called Hirschsprung-associated enterocolitis, which is inflammation of the colon, and is likely to be a consequence of this altered microbiome. Our work provides new mechanistic insight in what could be causing the inflammation."
The Eisen, Guillemin and Parthasarathy labs are part of the UO's Microbial Ecology and Theory of Animals Center for Systems Biology, which launched in September 2012 with a grant from the National Institutes of Health.
The NIH, National Science Foundation, Research Corporation for Science Advancement and Gordon and Betty Moore Foundation supported the research.
—By Jim Barlow, University Communications


